
UTAR
Centre for Research on
Communicable Diseases (CRCD), in collaboration with Centre for Corporate and Community Development
(CCCD), organised a webinar titled “Whole genome sequencing of bacterial
pathogens: Its potential and limitations” on 7 December 2020.
The
webinar was conducted via Zoom. It was moderated by Asst Prof Dr Saw Seow
Hoon from UTAR Faculty of Science and delivered by Prof Dr Yeo Chew Chieng
from Universiti Sultan Zainal Abidin (UniSZA) Medical Campus in Kuala
Terengganu.
In his
talk, Prof Yeo first gave a brief introduction on milestones in genome
sequencing and assembly. “In 1995, the first complete genome of a
free-living microorganism was published in Science. Sequenced using the
Sanger dideoxy method, this is what we now called ‘First generation’
sequencing technology, It is expensive, cumbersome and time-consuming.” He
then explained the two waves of sequencing technology innovation in the 21st
century—the second-generation (routinely termed as NGS) and the
third-generation sequencing (TGS). In order to provide participants with a
better understanding, Prof Yeo gave an idea of the differences between the
second and the third generation sequencing technologies. He further
explained the third generation sequencing technologies, focusing on PacBio
systems and Oxford Nanopore Sequencing.
Furthermore, he talked about metagenome sequencing, explaining that NGS and
TGS can be applied directly to clinical specimens—sequencing the DNA or RNA
extracted from patients sample by shotgun metagenome sequencing. He also
shared some drawbacks of the metagenomics NGS approach in clinical
microbiology, as well as the analysis about WGS of Acinetobacter baumannii
from Terengganu. He mentioned that the biggest challenge in introducing NGS
in clinical microbiology laboratory was the data analyses. He also
highlighted that dedicated and well-trained staff are required to ensure
successful implementation of WGS in routine clinical microbiology. Besides,
he also presented a simple cost analysis for Illumina sequencing and
nanopore sequencing, as well as discussed the current WGS issues.
“Sequencing technology, bioinformatics and informatics infrastructure are all rapidly evolving. The focus in these recent years is nanopore sequencing. The role of WGS and metagenome sequencing in clinical microbiology will increase in the coming years. This will transform clinical microbiology into a genome-powered and personalised diagnostic area,” he said. The talk then ended with a Q&A session.
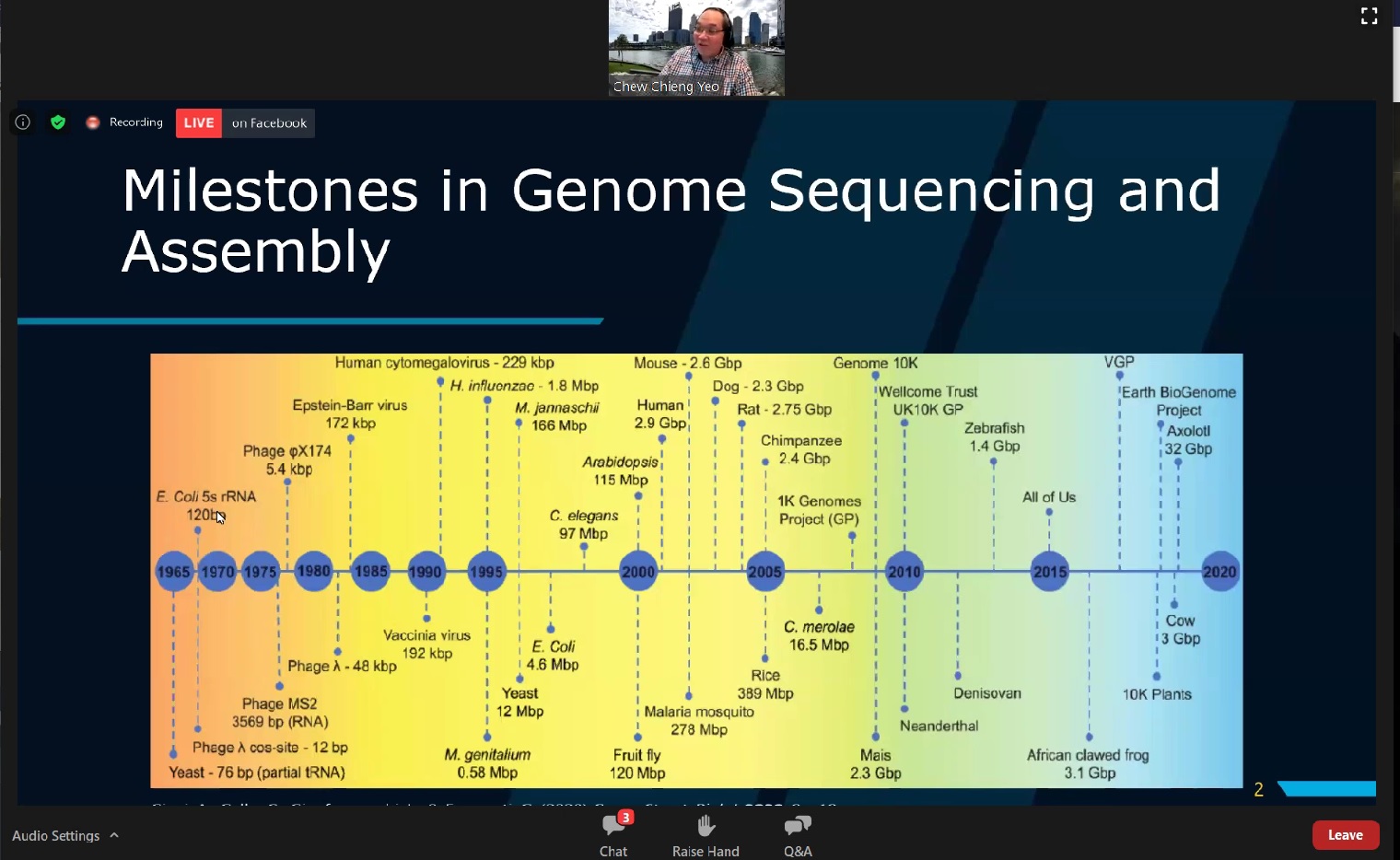
A timeline presented by Prof Yeo illustrating milestones in genome assembly
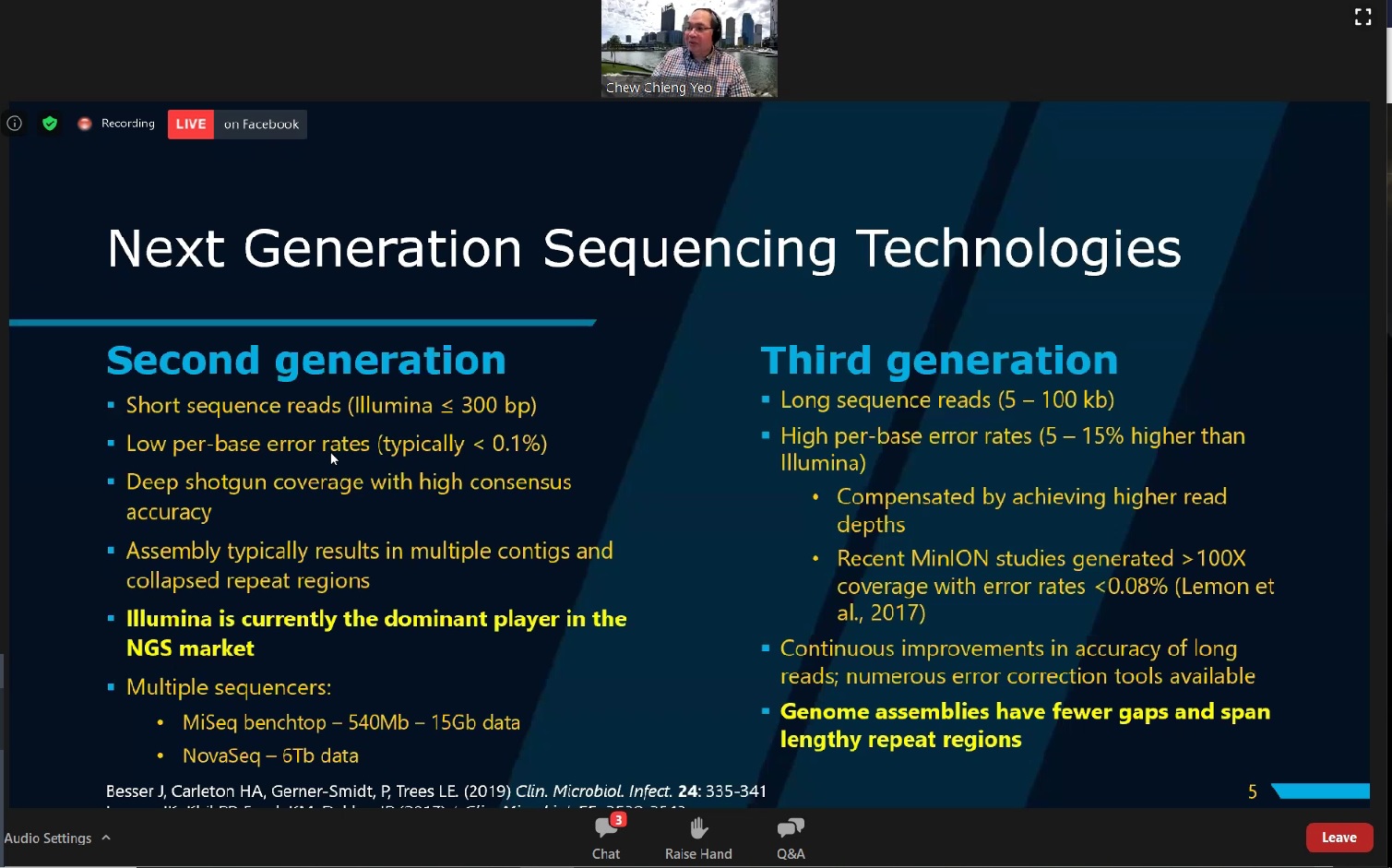
Prof Yeo presenting the differences between the second and the third generation sequencing technologies
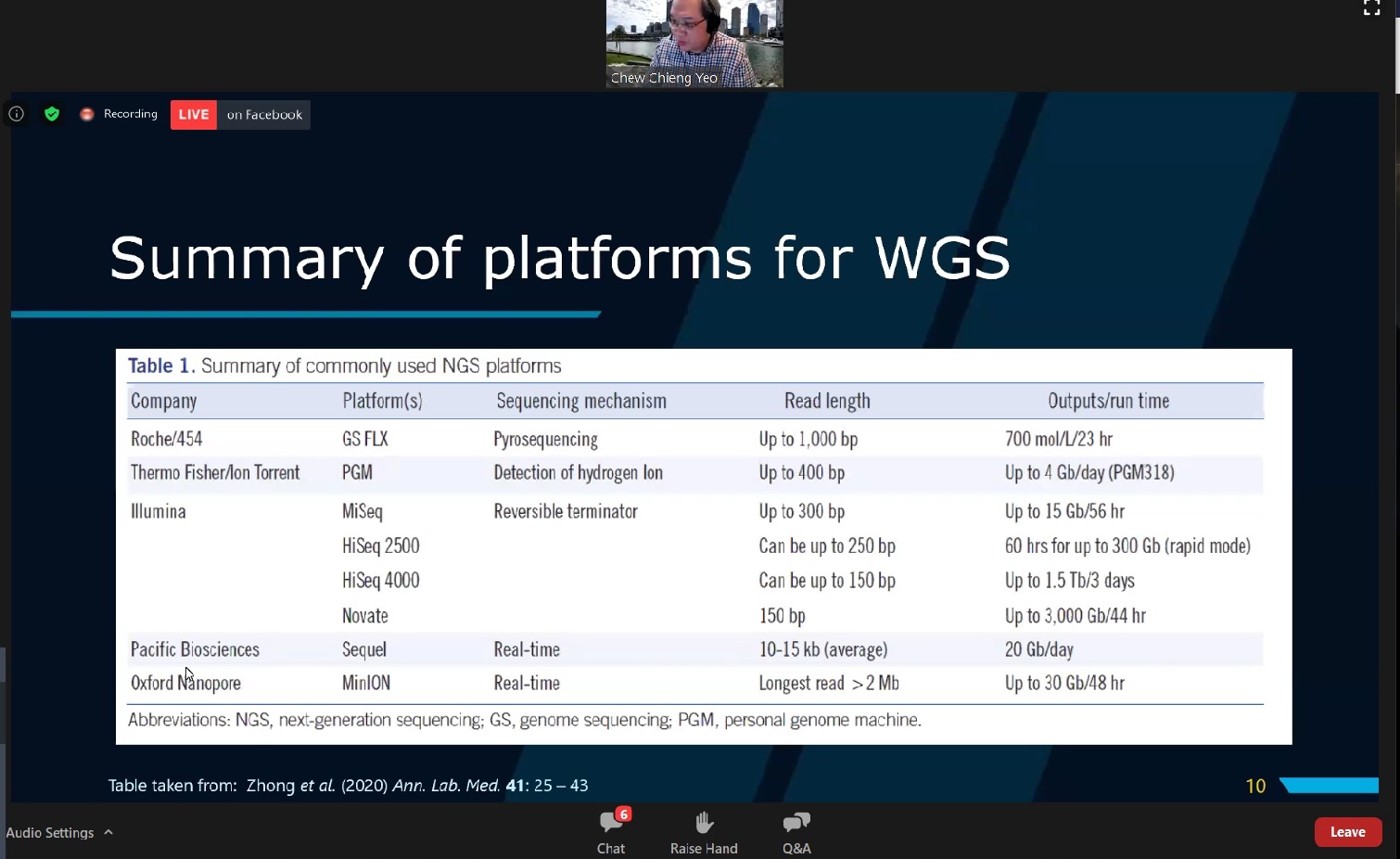
Prof Yeo sharing some platforms for WGS
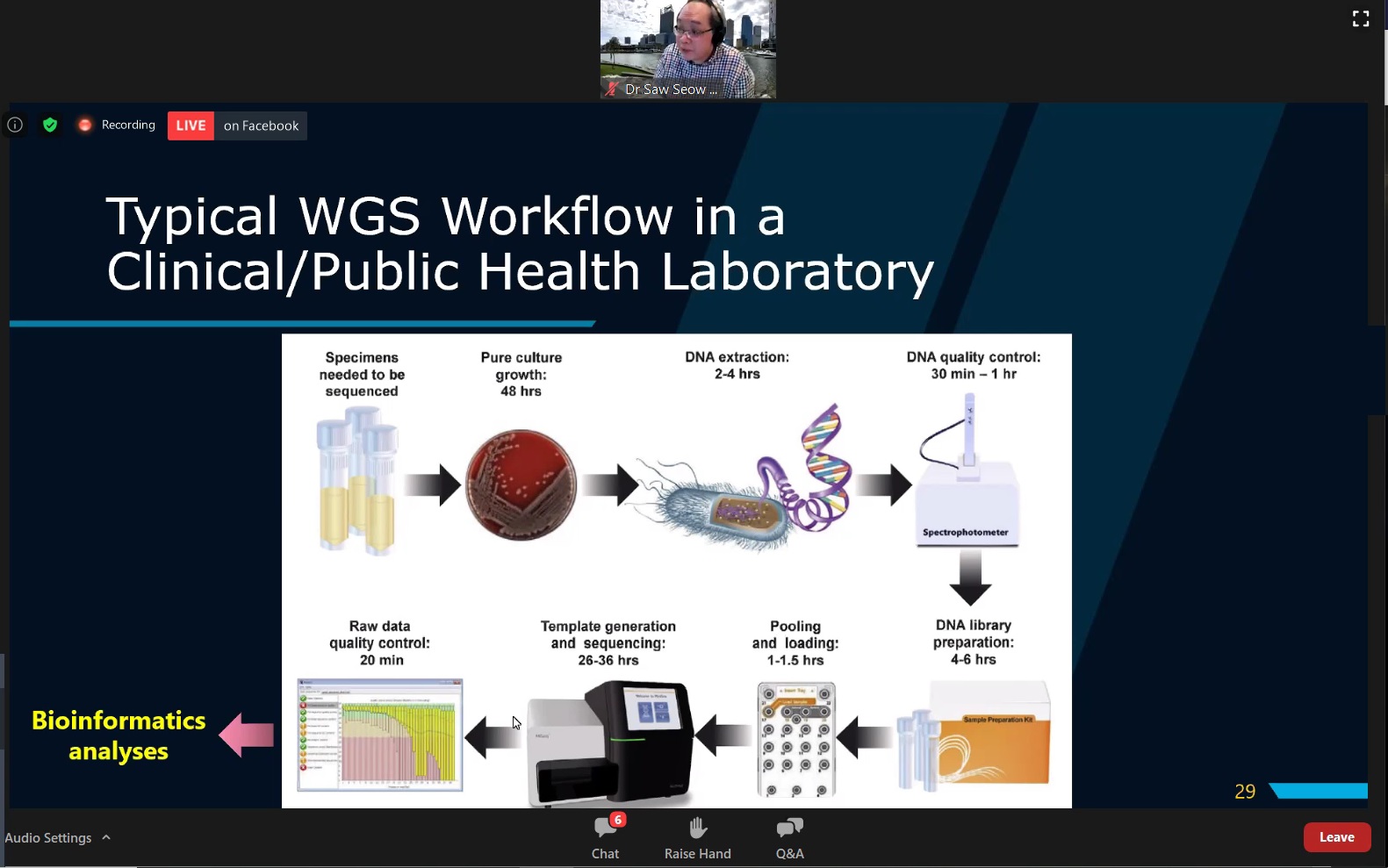
Prof Yeo presenting the typical WGS workflow in a clinical or a public health laboratory
Wholly owned by UTAR Education Foundation Co. No. 578227-M LEGAL STATEMENT TERM OF USAGE PRIVACY NOTICE